CALIBRATOR
- Methods
- Blending calibrated predictions with base predictions
Conditionally activate a calibration
Purpose
The Calibrator contains properties and methods for 1) validation of Variant diameter and height models against local observations and 2) auto-calibration of diameter and height models through simple linear regression of local observations.
All OSM models contain a property called CALIBRATOR which can be used to access and control CALIBRATOR settings (properties) and run calibration routines with OSM commands. The CALIBRATOR object can be accessed through the following command object hierarchy:
SIMULATION.MODEL.CALIBRATOR
…OR…
SIMULATION
MODEL
CALIBRATOR
Properties and Methods
Properties
|
Property |
Default Arguments |
Description |
|
MinObservations |
10 |
Minimum required tree observations per species for validation and recalibration. Must be > 0. |
|
MinSignificance |
0.05 |
Minimum statistical significance level (p-value) for validation and adjusting calibration. |
|
Mode |
MLR |
Calibration mode: · MLR = multiple linear regression (modified prediction = prediction + DBH + BAL + Stand BA) · MOD = simple modifier on existing predictive model (modified prediction = multiplier * prediction) |
|
Models |
DBHI,HT,HTI |
Models to try to calibrate: · DBHI = DBH increment · HTI = Height increment · HT = Height To turn on multiple models, state models separated by commas with no spaces following the Property; for example, ‘Models DBHI,HT,HTI’ would tell OSM to try and calibrate all models if any respective observations exist in the user’s tree list. |
Methods
|
Method |
Required Arguments |
|
Start Resets the Calibrator and begins
recording tree observations & predictions during one or more simulations. When calibrating, Tree Agitation is turned off. |
None |
|
Build [1] [2] [3] Performs model validation, recalibration,
and saves results to calibrationFile |
1) calibrationFile [String] Absolute or relative file path of calibration file. File extension determines file format and must be either: · *.osmcal – writes models in standard file format, or · *.osmc – writes models as AMEND commands. 2) appendStatistics [Boolean] If true, model statistics such as coefficients, variance explained, and p-values are appended to the file. 3) appendTrees [Boolean] If true, tree observations used to fit the models are appended. |
|
Load [1] [2] [3] [4] [5] Loads user calibrated species
models from an OSM calibration file (*.osmcal). Simulation cycles to apply
modified predictions in are controlled through arguments. Use Amend commands if you want to schedule
prediction modifications following an event or following onset of certain
stand conditions. |
1)
modelTypes [DBHI,HTI,HT] Model types to load from calibration file. See property Models for available types and usage. Multiple types can be stated if separated by commas with no spaces. 2) startCycle [Int32; must be ≥ 0] Simulation cycle to begin using modified predictions. 3) blendCycle [Int32; must be ≥ 0] Simulation cycle to begin blending modified predictions with base predictions. Modified predictions will be increasingly diluted linearly from 100% from blendCycle to 0% at endCycle. 4) endCycle [Int32; must be > 0] Simulation cycle to revert back to base predictions. 5) calibrationFile [String] Absolute or relative file path of calibration file. Only files with format extension ‘.osmcal’ can be loaded. |
|
LogPredictions [1] |
1)
Comma
separated output file to write tree-level information and predictions to. |
Build
a calibration file
The example below demonstrates a sequence of commands required to build a calibration file from a tree list with a subset of DBH increment measurements. For this example, assume individual tree diameter increment values are expressed as the average growth (cm/year) measured over the next 10 years.
First, set the data source. The tree list(s) contained in the data source must represent the before growth condition when calibrating increments. The tree list(s) must contain observations of tree height (HT), diameter increment (DBHI), or height increment (HTI) in order to calibrate those respective models. In this case, the OSM_TreeList table contains the DBHI column with DBH increments expressed as average growth (cm/year) measured over the next 10 years. If only a subset of trees in the plot have increment measurements, than that subset must be drawn in an unbiased way.
INPUTS.Source MyDatabase.mdb
Second,
set the simulation time horizon.
1) Total simulation years must equal the increment measurement period and be divisible by years per simulation cycle (YPC).
2) YPC should match the cycle length that will be used in calibrated simulations.
SIMULATION
YEARS 10
YPC 5
Ideally, YPC
should equal the increment measurement period; otherwise, some degree of bias
may be apparent between observed and predicted DBH values at the end of the
increment measurement period. If the desired simulation cycle length is 5, then
5-year increment cores should be used.
Third, optionally change calibration properties. Property values shown below are default settings and only need to be stated if you want to alter them.
SIMULATION
MODEL
CALIBRATOR
MinObservations 10
MinSignificance 0.05
Mode MLR
Models DBHI,HT,HTI
Forth, tell the Calibrator to start recording tree observations and predictions by model and by species during one or more stand simulations.
SIMULATION.MODEL.CALIBRATOR.Start
Fifth, simulate all stands in the data source through OSM. While simulating, the Calibrator will record (in memory) observed and predicted height, height increment, and diameter increment for each tree with observations.
SIMULATE
Finally, following all stand simulations, the BUILD command is called to:
1) Test for significant difference between observed and predicted values by species using a simultaneous F-test.
2) If significant difference is present, the procedure solves new linear models in a stepwise procedure as a function of base predictions, DBH, basal area of larger trees, and stand-level basal area. During the stepwise procedure, poorest predictors are dropped first and the model with the highest r2 value is selected.
3) If the revised species model is significant and outperforms the base model r2 value, then the model is accepted and written to the calibration file.
SIMULATION.MODEL.CALIBRATOR.Build MyCalibrationFile.osmcal True True
Two file formats can be written by the BUILD method. The extension used on the calibration file path (1st argument) determines file format and must be either:
- *.osmcal – writes models in standard file format, or
- *.osmc – writes models as AMEND commands.
In this case, standard format (*.osmcal) will be used. Standard format is required if you want to load the calibration into the model through the CALIBRATOR.LOAD method. Alternatively, command format (*.osmc) can be used to build AMEND commands for use in more complex simulations with conditional ACTIONS. An error will occur if the calibration file extension is not recognized.
The last two required arguments of the Build command tell OSM whether to append to the calibration file:
1) model statistics (e.g., error, p-value, coefficients) and
2) observations used to fit the models
Important
The CALIBRATOR limits species models to the range of tree DBH and basal area of larger trees (BAL) in respective species observations. These constraints can be manually adjusted in the calibration file by using a text editor (e.g., Notepad++). In addition, the Variant model may bound modified predictions if they are outside the range of maximum increments or maximum heights allowable for each species.
Load
a calibration file
The example below demonstrates a sequence of commands required to load the calibration file built in the example above.
First, if not already stated, set the data source of stands you wish to simulate with the modified calibration.
INPUTS.Source MyDatabase.mdb
Second,
set the simulation time horizon.
1) Total simulation years is up to you.
2) YPC should match the cycle length that was used to build the calibration file.
SIMULATION
YEARS 100
YPC 5
Finally, load the calibration and grow (SIMULATE) all stands in the data source through OSM.
SIMULATION.MODEL.CALIBRATOR.Load DBHI 0 3 5 MyCalibrationFile.osmcal
SIMULATE
All five arguments are required by the LOAD method. See LOAD argument descriptions in the Methods table above. The first argument (modelType) tells the Calibrator that only certain model types should be loaded from the calibration file. This can be useful for setting different start, blend, and end cycles for different model types, or if using different calibration files for different models. Model types are managed independently within the Calibrator, so loading DBHI models will not clear HTI models previously loaded. To load all models from a calibration file, append all model type codes into the first argument separated by commas (no spaces permitted); e.g., “DBHI,HT,HTI”
Blend calibrated predictions with base predictions
LOAD command arguments 2-4 specify rules about when and how calibrated models should be used during a simulation:
2) startCycle specifies the simulation cycle that the calibrated model should be activated in.
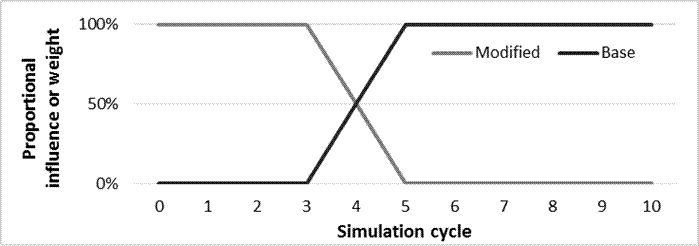
3) blendCycle specifies the cycle to begin blending calibrated predictions with base predictions. Modified predictions will be increasingly diluted linearly from 100% from blendCycle to 0% at endCycle.
4) endCycle specifies the simulation cycle to revert back to base predictions.
For the example above, the calibration will be activated immediately (cycle 0) and persist until cycle 3 (15 years). For cycles 3, 4, and 5, predictions from the calibrated model will be linearly diluted with base predictions starting at 100% calibrated predictions in cycle 3 down to 0% by cycle 5, respectively. From cycle 5 on, only base predictions will be used.
Clear a calibration
Note that once a calibration has been loaded, it will remain in effect until:
1) The OSM program exits.
2) Calibrator.Start is called.
3) Calibrator.Load is called to replace a specific model(s).
4) Calibration.Clear is called.
Conditionally activate a
calibration
In some cases, users may wish to modify predictions under certain situations:
· Following one or more partial cut entries, where timing of each harvest entry depends on stand structure.
· Following natural disturbance, where timing of disturbance is dependent on stand structure.
The LOAD method cannot schedule activation of a calibrated model following stand treatments or disturbances if timing of those events is conditional on stand composition and structure. The LOAD method is also limited to a single simulation cycle window when it can be applied.
To enable conditional calibration activation, calibrated models must be stated as AMEND commands within the scope of an ACTIONS definition, which can state under what stand conditions or time period following treatment or disturbance the calibration should be activated.
The command syntax to represent calibrated models as AMEND commands is not straightforward; however, by switching the calibration file extension from *.osmcal to *.osmc when building a calibration file (see BUILD command), OSM will automatically write out the required commands to implement the calibrated model as AMEND commands, as shown below. These commands can then be manually copied from the calibration file into your ACTION definition.
In the example action below, a 33% basal area removal will be scheduled as soon as the stand contains 24 m2/ha of basal area. When activated, diameter increment (DBHI) will be modified for white birch, yellow birch, and white spruce according to the respective species models shown below (generated from the CALIBRATOR.BUILD command).
Note:
· ‘AMEND 0..5’ states modifications should be applied between cycles 0 and 5 relative to the cycle treated.
· ‘BLEND 2 5’ states modified predictions should begin blending with base predictions between three cycles after treatment (100% modified predictions) and five cycles after treatment (100% base predictions). This is analogous in most ways to blending calibrated predictions with base predictions described previously for the LOAD command.
· The modifications are limited to certain species and DBH and BAL ranges. These ranges reflect the extent of observations used to fit each species model and can be adjusted manually.
ACTIONS
def PartialCut
OPERABLE
BA >= 24
CUT 0
Retention
BA >= 66%
LOCK 5
AMEND 0..5
BLEND 3 5
DBHI = "0.5018752-0.01084047*StandBA" in BEPA with DBH 4.04 39.34 BAL 0 20.73
DBHI = "0.259392+0.5510734*DBHI-0.009267476*StandBA" in BEAL2 with DBH 1.16 57.52 BAL 0 29.5
DBHI = "0.9103431-0.02245477*StandBA" in PIGL with DBH 1.01 38.34 BAL 0 15.93
See AMEND command help on “using equations in modifier statements” to better understand the last three lines above.
Log tree-level predictions
OSM tree
predictions can now be output directly to a comma separated text file without
having to run statistics and generate calibration files. Other useful stand and
tree information is sent to this file. This is useful for more closely
examining, graphing OSM predictions against observations or to other models.
Predictions can be joined with your original tree list table using SurveyID and TreeID columns. Only
tree records with observations of DBHI, HT, or HTI will be written to file. Use
‘Models’ command to specify which observation types should be written to file,
or ignored.
SIMULATION
MODEL
CALIBRATOR
Models DBHI,HT,HTI
Start
LogPredictions
MyFileName.csv
SIMULATE
SIMULATION.MODEL.CALIBRATOR.Clear